
Exercise 1: De Novo Assembly using Spadesĭe novo assembly is one the most computationally demanding processes in bioinformatics. In conclusion, the de Bruijn graph assemblers are more appropriate for largeĪmounts of data from high-coverage sequencing and have become the programs ofĬhoice when processing short reads produced by Illumina and other established

Waterman, 2001), Velvet (Zerbino & Birney, 2008), ABySS (Simpson et al., 2009),ĪllPaths (Butler et al., 2008) and SOAPdenovo (Li et al., 2010). Programs implement de Bruijn graph algorithms, including Euler (Pevzner, Tang, & Stored at most once regardless of how many times it occurs in the reads. Is especially suitable to handle the large number of reads because each k-mer is Is resolved by finding a path that visits every edge in the graph. Since edges correspond to all k-mers existing in the sampled genome, the assembly In the figure above the sequenceĪTGGCGTGCA with 3-mers and overlap of 2 base pairs (bp). Therefore, groups of overlapping reads are not actually computedīut rather represented as paths in the graph. Represents an observed k-mer and its adjacent nodes the prefix and suffix of the Overlapping sub-reads, and then use the latter to build the graph. Modern de novo assemblers work (Compeau, 2007).ĭe Bruijn graph assemblers start by splitting the set of reads into k-mers, a set of De Bruijn graph assemblers are the state-of-the-artĪpproach for data sets composed of many thousands of short reads.Īn example de Bruijn graph – the graph on the left is how These two algorithms (greedy and OLC) are more suited to fewer, longer reads than Paths along the graph show likely contigs. It requires a very time-consuming first step, where all reads are comparedĪgainst each other. OLC assemblers build an overlap graph, in which nodes represent the reads and edges the (Warren, Sutton, Jones, & Holt, 2007), the first short-read assembler, is based on thisĪpproach as well as its two descendants SHARCGS (Dohm, Lottaz, Borodina, & Himmelbauer, Length or the percentage of identity between reads along their joining region. Such scores are normally measured as the overlap The greedy algorithms consist of progressively adding single reads into contigs byĮnd-to-end overlapping, starting with those reads with the highest overlap score andĮnding once no more joins can be found. Unidirectional which means that they can only be traversed in the direction of the arrow.įor example paths in this graph include 3-2-5-1, 3-4-6 and 3-2-1. In the graph to the left there are 6 nodes and 7 edges. Graphs are sets of points called ‘vertices’ or ‘nodes’ joined by lines called Graph theory is a branch of discrete mathematics that studies problems Nodes and edges, being classified into three main groups: greedy, Overlap/Layout/Consensus (OLC)
bacterial) with relatively low fragmentation.Ĭurrent assemblers employ graph theory to represent sequences and their overlaps as a set of To obtain assemblies from small genomes (e.g. 100 read depth, >150bp – Illumina HiSeq2500) and paired-end information make it feasible Overlaps and false positives (Miller, Koren, & Sutton, 2010)ĭespite all mentioned limitations, the high coverage currently achieved, growing read lengths Mismatches when overlapping reads and joining regions, possibly leading to discarding true Sequencing errors add difficulty since algorithms must allow certain Repeats greater than the read length by employing their expected separation and orientation as These are two reads that are separated by a gap of known size. In addition, the modern hardware commonly output paired-end reads. Greater the chance of observing overlaps among reads to create larger contigs and being able to the number of reads over each part of the genome. These issues can also be overcome by increasing the coverage depth, Longer read lengths will overcome these limitationsīut this is technology limited. Is lower and repeat regions harder to resolve. Read lengths are short and therefore detectable overlap between reads However de novo assembly from next generation sequence (NGS) dataįaces several challenges.
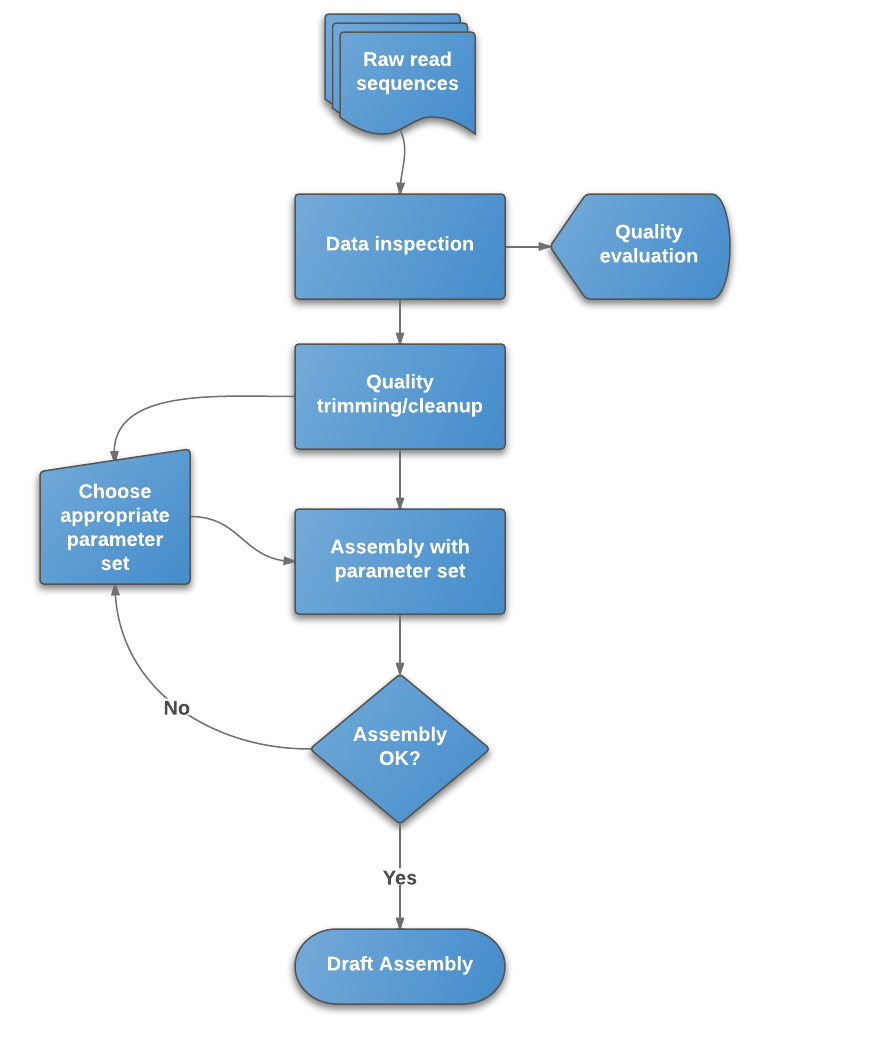
Contigs are joined into scaffolds covering, ideally, the whole of eachĬhromosome in the organism.

It follows a bottom-up strategy by which reads are overlapped and We have already explored mapping, where the sequence data is aligned to a known reference genome.Ī complementary technique, where no reference is used or available, is called de novo assembly.ĭe novo assembly is the process of reconstruction of the sample genome sequence withoutĬomparison to other genomes.
#De novo assembly geneious tutorial how to
The main challenge of sequencing a genome is determining how to arrange reads into chromosomes. Example fallback content: This browser does not support PDFs.


 0 kommentar(er)
0 kommentar(er)
